m6A is a methylation modification found in thousands of RNAs in cells which affects different aspects of RNA regulation—its stability, how it’s processed, and how much protein it produces. It is a fundamental component of gene expression that is critical for the function of healthy cells and contributes to human disease when it is misregulated. For example, too much of the enzyme that deposits m6A in RNAs has been linked to leukemia and other cancers. Now, a published paper from the Meyer Lab in Molecular Cell sheds new light on several key features of m6A biology that have been missed by other studies.
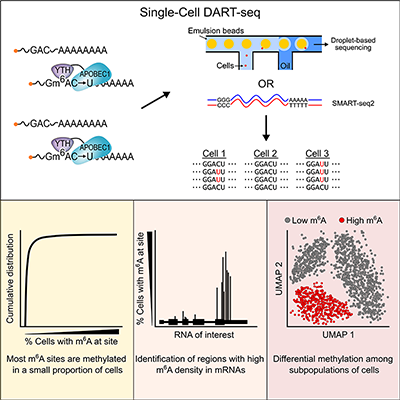
Although discovered back in the 1970s, much of the progress in understanding m6A has been made in the last decade with the development of high-throughput sequencing-based approaches for identifying sites of methylation within cellular RNAs. However, these methods require large amounts of RNA and have therefore been limited to looking at thousands or millions of cells at once. The new study from the Meyer Lab takes a completely different approach by using a novel m6A detection technology that enables the identification of m6A sites in single cells. The method provides unprecedented insights into how m6A is regulated.
Researchers have wanted to use single-cell sequencing to look at m6A, but achieving that level of resolution has been a major challenge. However, Matt Tegowski, PhD, a postdoc in the Meyer Lab and first author on the new paper, was undeterred. Adapting a novel m6A detection method previously established in the lab, Matt first attempted to identify m6A sites in a few single cells to see if the approach would work. After achieving initial success, he went full force and profiled m6A in thousands of individual cells—essentially improving on existing technology by several orders of magnitude.
Matt found that RNAs have many more methylation sites than previously thought and that most of these sites occur in a small percentage of cells throughout the population. He also discovered that groups of cells can be characterized by their methylation patterns, which might provide a new way to classify subtypes of cells in the future. These discoveries pave the way for future studies examining RNA methylation within individual cells that make up complex tissues, such as the brain, heart, or even a tumor sample. Matt’s technique will answer so many questions.
What does this do for disease therapies and even cures? So far, researchers’ ability to examine m6A within an individual diseased cell has not been possible, and their understanding of how m6A contributes to pathogenic states can therefore be clouded by nearby healthy cells. This new technology now enables a whole new level of understanding RNA methylation, reshapes the way we think about m6A regulation in cells, and paves the way for future work painting a clearer picture of how m6A contributes to human disease.